The Century of Biology: Introduction
For starters, what is biology? It’s the study of all living things. Every living being on earth is the triumphant display of biological processes at work. Every man, woman, and child; every bird, bacteria, and leaf; and every cell within. It’s the study of life, growth, and death; of proper cellular functioning and disease. Before humans, biology was the only source of food, medicine, shelter, energy, etc.
It is the engineering platform devised by Mother Earth to build the physical world around us, and it’s the most precise and efficient engineering platform ever created, many orders of magnitude superior to that of humanity’s. She can program at the accuracy of atoms: the placement of atoms determines the genome sequence coding for organisms; proteins nearly one-one billionth of a meter across are the machines driving all the physical functioning of all living things; cells one-millionth of a meter across interact to form tissue which form organisms. Her efficiency is demonstrated by the fact that she doesn’t generate a single atom of unused waste. The process is self-renewing and self-perpetuating at all scales. Organisms interact to find a continuous, dynamic equilibrium of life and death. When cogs in the wheel break down and die, they are decomposed into individual molecules to form the matter for the next being. A dead tree get digested by microbes into soil which in combination with the sun provides the nutrients for plants to grow which in turn feed all animals which feed on each other. That metabolic process of eating provides the energy to live and to mate which produces new life. That life begins at first in the form of a single cell which itself divides trillions of times to form full sized organisms. Moreover, all this happens autonomously. There’s no floor manager overseeing the machinery at work, on / off buttons, lunch breaks, Christmas bonuses, or thank you’s. It happens without any of us thinking about it.
How does this all work under the hood? To power the circular manufacturing platform described above, nature created an information processing system in the physical world. While humanity’s information processing systems – digital devices and networks – rely on physical phenomena like moving around electrons to perform the underlying computing, these are only a physical means to a digital end. Electrons are flowing around to virtually represent an Angry Bird hurling towards a pile of blocks on your phone. Nature, at its most fundamental level, also manipulates physical and chemical processes to make things happen. For instance, pulses of electricity moving across chemical gradients1 underlie your brain’s computing, the beating of your heart, the information transmission from your finger to your brain that the bowl of ramen your touching is scalding hot. You turn that ramen into energy by a chemical cycle that involves ripping apart hydrogen bonds and harvesting the energy released1. Moreover, chemicals are the primary way that cells communicate with one another.1 The ultimate end goal of nature, however, is not to move around bits, but atoms.
Two other fundamental differences between humanity’s digital information processing systems and nature’s physical ones are their respective complexities and scales. Interdependent networks of genes, molecules, and cells controls development (cell division, differentiation, death) and responsiveness to environment (information measurement, computation, reaction).
Consider for a second that every living organism on the planet starts its journey as a single cell. How do us humans go from the complexity and scale of single-celled bacteria to a collection of 38 trillion cells? Moreover, how does that one template cell differentiate into hundreds of unique types, each with specific jobs and molecular constituents so that we aren’t an amorphous blob of uniform cells but a body with distinct organs, including those that can sense sound waves and convert them into electrical signals, those that detect and attack invading pathogens, and those that endow us with not just the ability to make sense of what you’re reading but to have self-reflective consciousness?
The answer starts with our genetic code, which serves as the blueprint for our development. Our genome, 3 billion letters long, codes for 20,000 genes and each gene codes for a specific action to take place. That action may be the production of a protein, like the one that embed in the membrane of our ears’ hair cells and help facilitate our hearing. Or the gene may trigger a cascade of actions, like those involved in cell division.
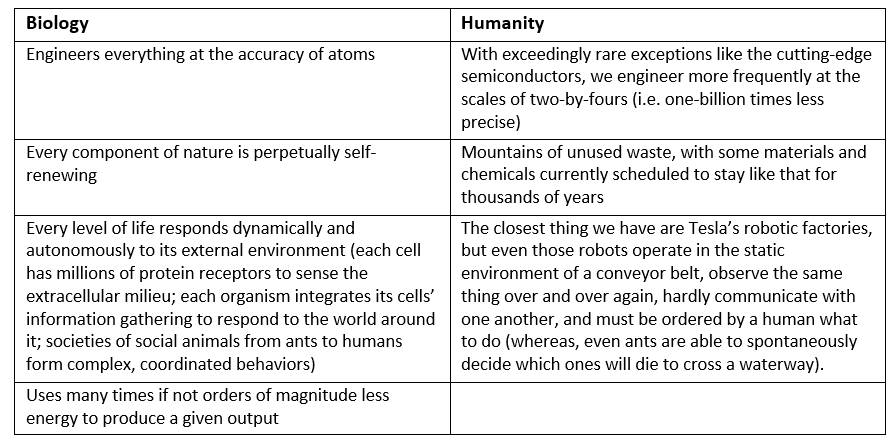
Indeed, genes may encode for the turning on or off of other genes. Our genetic code thus forms a network of genes in which one triggers another which combines with a separate one to trigger the activation of a certain set of other genes. This gene regulatory network is structured hierarchically but with interdependencies at every level, similar to a modern corporation. The first couple levels or units, to continue the corporation analogy, are 1) global genes that use wide-scope directives that engage 2) cross-functional teams of genes to coordinate the actions of 3) general managers of modular department-level networks that ultimately run 4) particular functions. A final layer are inter-departmental genes that facilitate crosstalk between modules and integrate those signals to achieve a coherent response. [1] To visualize these networks, consider the graphs below which provide two approaches to conceptualizing the gene regulatory network (GRN) of the bacteria E. coli. The one on the left maps it like a corporate hierarchy chart, the right as a node network.
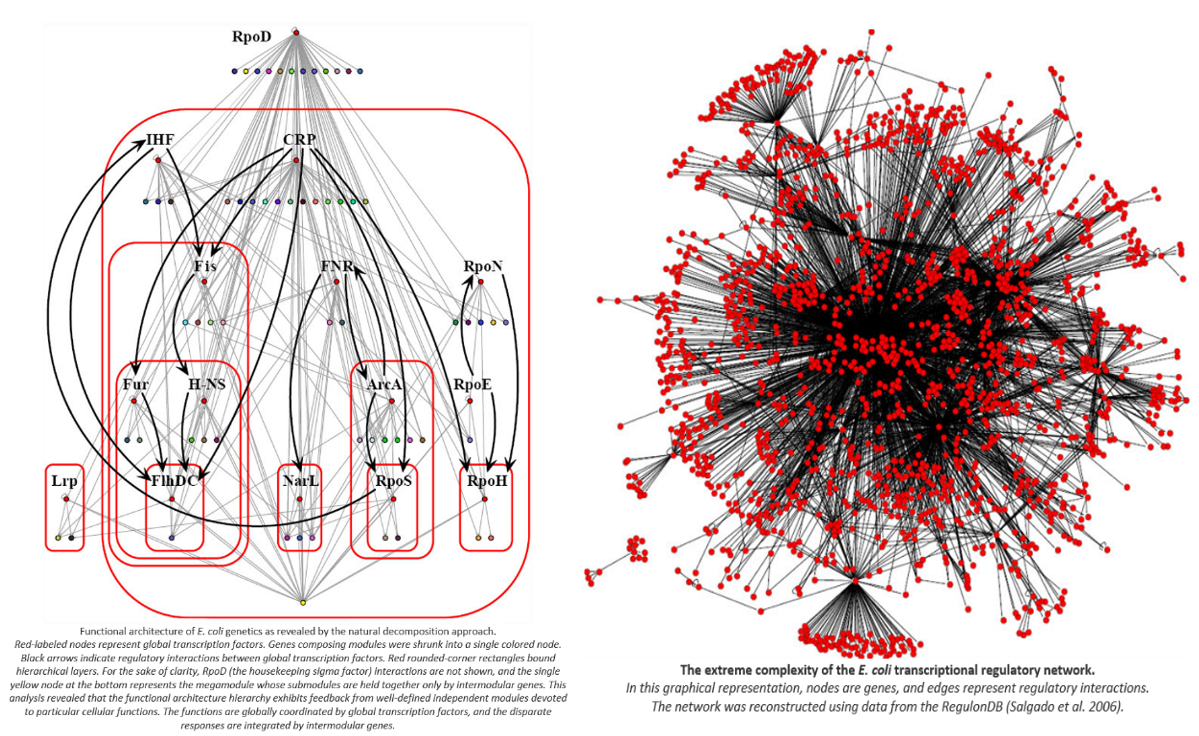
In this sense, the genome works not like a book in which you sequentially read one page after another, but more like electrical circuits where certain genes turn on or off networks of other genes which feed forward into the activation of other genes. Gene circuitry can include Boolean logic (e.g. only activate gene x if both gene y and z are activate) as well as other input-output functions like IF-THEN statements. The diagram below shows a stylized example of a multistep circuit:
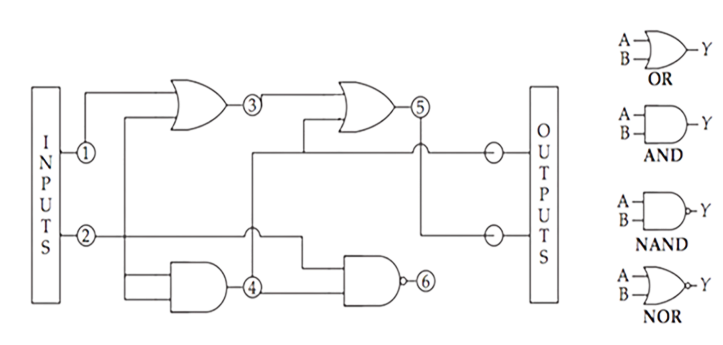
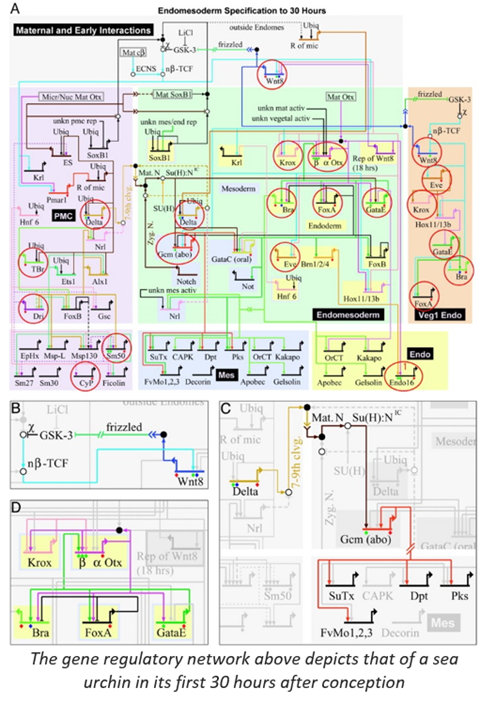
In real life biology, the circuitry is unimaginably more complex. The series of circuits orchestrates the process of organism development, causing specific genes to be on or off in any given cell over space and time. The graph below shows the GRN circuitry for a sea urchin in its first 30 hours after conception. Extrapolate the complexity of the diagram for such a simple organism to imagine the complete developmental GRN for a human over its 750K hour lifetime from the single cell conception to 38 trillion cells at physiological maturity to eventual death.
I began this discussion of GRNs by saying the answer merely starts with the genome because every cell, whether it’s a lung cell, a neuron, or one in your eyeball has the exact same genome in its nucleus. Only a small subsection of genes is turned on in a given cell at a given time, thereby determining its type (whether it’s a lung cell, neuron, etc.), its stage in the progression from cell division to death, among other cell identities. Moreover, the sequences of letters that makes up the genome only scratches the surface of determining which genes are turned off or on. DNA is constantly physically and chemically modified to make certain genes more or less likely to be activated.[2]
The complexity of GRNs devoted to development is compounded by the fact that the organisms must constantly be adapting and responding to its external environment. To do so, the genome also encodes for the construction of physical regulatory networks at all levels of the organism, from between individual molecules like proteins to cells to organs. Proteins and other receptors embedded in the cell membrane sense for the cue they’re designed to detect. That signal gets relayed to its proper endpoint via chemical particles acting as messengers, chemical or physical modifications of macromolecules, or physical action performed by macromolecules (movement, an enzyme breaking down another object, etc.).
As an example, consider the graph below depicting a long cascade of reactions starting with an adrenaline receptor sensing an adrenaline molecule and ending with a gene being turned on. The activation of the adrenaline receptor activates its associated G-protein, which in turn awakens a membrane-bound enzyme called adenylyl cyclase. Its gears start churning and pump out AMP molecules, a second messenger small molecule. Such molecules can amplify the initial signal hundreds or even thousands of times to make sure the rest of the cell gets the message, so to speak. The AMP signal activates protein kinases (PKA, in the diagram), which then makes a chemical modification called phosphorylation to multiple protein substrates by attaching phosphate groups to them. The PKA enters the nucleus, recruits the proper transcription factors, which causes the transcription of the desired gene at the promoter site. Once each of these molecules have done their job dutifully, they’re degraded by enzymes.
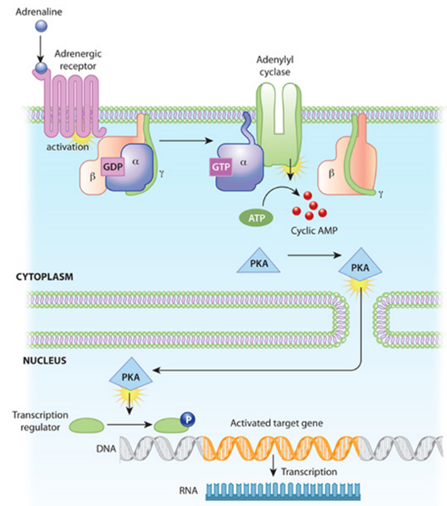
In addition to triggering long cascade of consecutive actions, these circuits can execute sophisticated logic, akin to input-output functions encoded in the genetic circuitry. They can perform Boolean logic, amplification or Sigmoidal activation, positive or negative feedback loops, etc. Many of the possible circuits are detailed in the table below.
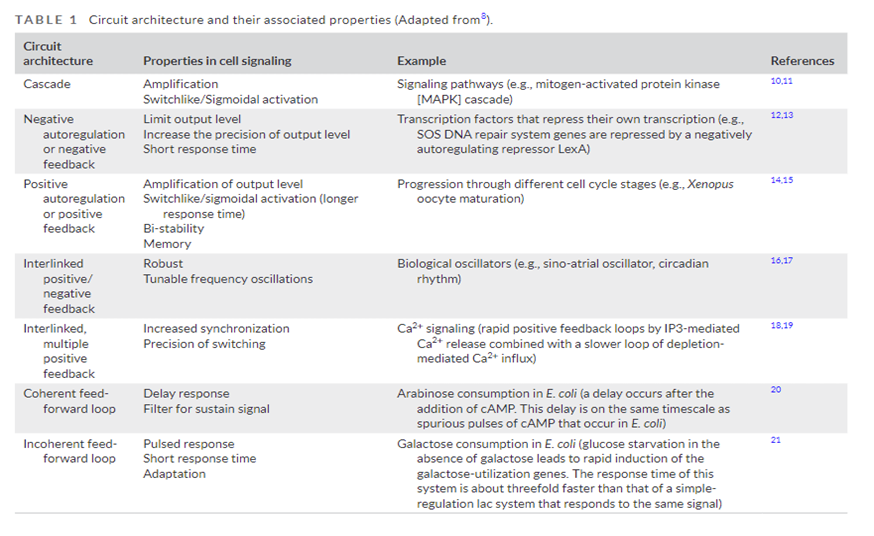
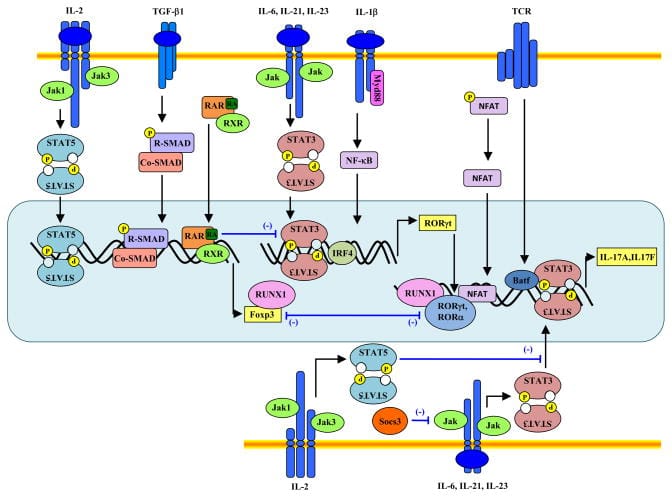
These circuits are all modular. So, they can be used together in any combination desired, enabling a massive combinatorial space of possible ways to control cell function. As an illustration of this, consider that immune cells have two receptors that must both be activated (an AND gate) in order for the cell to turn on; the intensity of the signal received must meet a certain threshold for it to continue (Sigmoidal activation). These two features make sure the immune cell is highly selective in which cells in targets and doesn’t attack our own body’s healthy cells. If those two requirements are met, then a positive feedback loop of signal amplification is initiated to make sure the cell fully activates to respond to the threat (IF-THEN and Amplification). Once activated, several signaling pathways consummate in the importation of transcription factors into the nucleus to initiate the expression of genes involved with cell proliferation and differentiation. An army of one rapidly becomes full platoon of immune cells and begins waging war against the pathogen. Additionally, at each node in this multi-step function, negative feedback loops are in place to prevent the overactivation of immune cells which can have detrimental effects. Here's a representation of the actual circuits involved with the activation and differentiation of an immune cell:
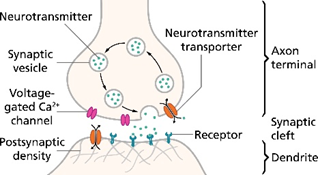
These networks of physical circuits also enable our cells to communicate with one another. The intercellular signals are predominantly transmitted via chemicals and electricity. These messages can be sent over various regulatory scales and physical distances. A cell can talk to just its direct neighbor (e.g. neurons talking to each other as seen in the diagram on the right). Or, that inter-cellular gossip can be replicated over the course of the whole body such that skin cells in your finger can transmit the sensation of represented in an electrical signal through all the cells leading up to your brain, like a massive game of telephone. Cellular communication can be harnessed for regulation of the body at both local and global scales. For instance, the GRN can trigger the release of growth factors into the blood stream to spur cell differentiation and growth throughout the body.
Alright, now that we have a sense for how much big the combinatorial space is for the programming of our genetic and physical circuitry, it’s time to get a sense for the scales these circuits operate on. Each of our cells contains tens to hundreds of millions of proteins[3] and tens of millions of receptor proteins are in each cell’s membrane sensing the external environment[4]. Thus, an immense amount of computation about how to properly respond is continuously being undertaken in each cell. These reactionary circuits combined with the GRN controlling cell development engender thousands of genes being activated in every cell every second1. Each human has 38 trillion cells.
Moreover, these regulatory networks are present across of lifeforms. The machinery is silently whirling away, powering the underlying processes of everything in the living world. When these circuits are functioning properly, the organism’s cells are healthy. When they fail, disease sets in. Biology is the study of these circuits across all living organisms. It’s about understanding the greatest information processing system ever devised.
And, we now finally have the tools to measure biology’s circuitry. In the last two decades alone, we’ve developed the capacity to do the following, all at scale:
- Read, write, and edit the code of life (DNA)
- Used this to understand what genes are being expressed in cells
- Visualize the theatre of life at every possible resolution, from making out individual atoms to mapping entire organs
- Collect multimodal data from living patients at population-sized cohorts via continuous health monitoring devices like wearables and advanced diagnostic tests
- Simulate human biology in the lab with miniaturized organ replicas
Meanwhile, the dramatic progress in artificial intelligence and other advances computing methods have increasingly allowed us to make sense of the massive amount of data generated from the tools mentioned above and will be indispensable to teasing out biology’s complexity. Indeed, we’ve already seen that progress in AI broadly be ported over to biology, with hundreds of foundational models being developed in the space.
All of these techniques have improved exponentially since their invention across speed, cost, and scale. Many have outpaced Moore’s Law. Our understanding of biology will track this progress, because more so than in any other discipline, tools are the rate-limiting factor in biology. Scientists can only understand what they can measure. We didn’t know about the existence of cells until the invention of microscopes, DNA’s double helix until x-ray crystallography, the scope of genetic diseases until next-generation DNA sequencing, etc.
Part I of this essay delves into the tools we’ve developed to understand biology, detailing the trajectory of each tool’s progress, where it currently stands, what it allows us to do, and where it may be headed in the future.
Part II of this essay describes our progress in programming, controlling, and creating biology. Just as basic science tools fuel our understanding of biology, our understanding of biology powers our ability to control it to our desired ends.
At our current juncture, there are eye-popping gaps between the current practices of say our healthcare systems, what scientists and entrepreneurs are already proving possible, and where a reasonable extrapolation of the future could take us. Doctor’s offices look exactly like they did 50 years ago. Annual checkups, the closest thing we have to continuous health monitoring, still use a point-in-time blood pressure measurement and a rock tapped against our knee as the state-of-the-art technologies. We still diagnose health and treat disease based on population averages rather the specifics of individuals. And, we still have no way of combatting the diseases we are all most likely to die from (old age, heart disease, cancer, Alzheimer’s, diabetes). I can’t help but have the feeling that our understanding of and ability to deal with disease is like that of the Dark Ages compared to where it will be in the not-too-distant future.
As the reader will see over the course of this essay, that future is in many ways already here, it’s just not evenly distributed. Here are some examples of current realities feeling more akin to science fiction:
- The COVID vaccine works by reprogramming our cells’ circuitry by giving them the genetic instructions they need to mass manufacture the antibodies to fight the virus. Moreover, consider that we sequenced the DNA of the coronavirus in days, used that to prototype conceivable drug designs within weeks, in under a year had a product with 90%+ efficacy and tremendous safety characteristics was rolled out globally, and to date nearly 10 billion doses have been administered at the cost of a Chipotle bowl apiece. This would’ve taken many years if not over a decade to do under prior technological regimes.
- This year, we first directly edited the genome of sickle cell anemia patients at individual base pair resolution to permanently cure them of the illness. Gene therapies have also been used to cure a hereditary blindness, permanently lower cholesterol, etc.
- Wearables, powered by seven decades of Moore’s Law, are now capable of continuously gathering clinical-grade health data from heart rate to blood oxygen to glucose levels to sleep. Soon, that will be augmented by many more types of data about our physiology from many of the devices we interact with intimately in our daily lives from our bed to clothing to toothbrush to toilet. They will analyze our microbiome, screen us continuously for all kinds of cancers as well as less serious illnesses, etc.
- Ten years ago, our understanding of transcription factors led to scientists figuring out how to use them to turn back the age of cells to zero. Now companies are looking to systematically investigate the combinatorial space of all transcription factors to find therapies to reverse the aging of our immune system (which helps explain why cancer rates increase exponentially with age) as well as companies looking to use transcription factors to industrialize the production of differentiated cells for research purposes.
- Ten years ago, we first cured a cancer patient by extracting their immune cells from their body, re-engineering the cell’s circuitry by attaching a receptor from another cell in the body that makes the cells selectively attack cancer cells (which the immune cells had previously ignored). We’re now adding complexity to that initial circuit by implementing IF-THEN statements, Boolean logic, etc. in these engineered immune cells to increase their specificity, safety, and survival.
- Scientists have even begun exploring making inter-cellular circuits such that different cell types, engineered for specific jobs, team up together to combat the most complex of diseases.
With the advent of RNA, gene, cell, and even inter-cellular therapies like those mentioned above, we now have the primitives necessary to cure effectively all human diseases, at least in theory. And yet, human pathology is only the tip of the iceberg for biotech. Just as biology is the study of all living beings, biotech is the application of that understanding to use biological circuitry to make just about anything in the physical world. Scientists and entrepreneurs are engineering gene circuits in bacteria and plants as well as cell-free enzymatic processes to produce everything from drugs to food to plastics to t-shirts. As some examples:
- Impossible Foods identified the gene encoding for the production of heme (the molecule that gives meat the bloody, irony taste) in plants, transduced it into the DNA of bacteria, and them produce the molecule at scale. Other companies are controlling the differentiation of animal stem cells to grow meat in vats that’s molecularly identical to that derived from animal carcasses. Still other companies are engineering bacteria to convert greenhouse gas emissions into protein-rich foods.
- Researchers devised the following gene circuits in yeast to manufacture a medicinally valuable chemical that acts as a neurotransmitter inhibitor:
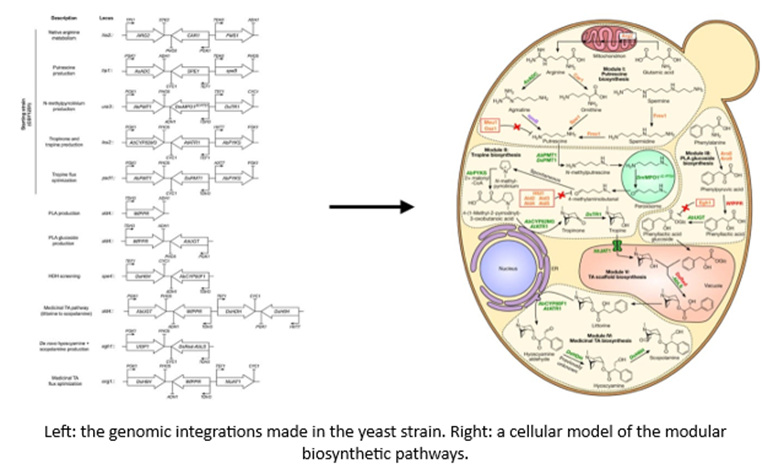
- A team recently transduced spiders’ genes coding for spider silk into regular silkworms to make a new type of fiber that has a combined strength and stretch unparalleled by any current materials.
- Other groups are making sustainable fuels with enzymes; plastic out of biomass, emissions, or waste; and biological processes to break down the plastic already in existence into useful components.
In summary, only just now do we have the tools necessary to understand and then control biology. We’ll use that capability to mitigate and cure complex diseases like cancer, Alzheimer's, even aging itself; grow food to feed 10 billion people without slaughtering ever-more animals; create a circular economy for plastics, fuels, chemicals, clothing; and more – all while helping humanity hit its climate objectives.

Read next section: Reading, Writing, and Editing Life's Code
[1] For more detail on the hierarchical structure of GRNs, see this paper
[2] Epigenetic mechanisms, including DNA methylation and imprinting, noncoding RNA, post-translational modifications, and other mechanisms, further enrich the cellular portfolio of gene expression control activities. Even when transcription factors are present in a cell, transcription does not always occur, because often the TFs cannot reach their target sequences. The association of the DNA molecule with proteins is the first step in its silencing. The associated DNA and histone proteins are collectively called chromatin; the complex is tightly bonded by attraction of the negatively charged DNA to the positively charged histones (Table 1). The state of chromatin can limit access of transcription factors and RNA polymerase to DNA promoters, contributing to the restrictive ground state of gene expression. In order for gene transcription to occur, the chromatin structure must be unwound.
Chromatin structure contributes to the varying levels of complexity in gene regulation. It allows simultaneous regulation of functionally or structurally related genes that tend to be present in widely spaced clusters or domains on eukaryotic DNA (Sproul et al., 2005). Interactions of chromatin with activators and repressors can result in domains of chromatin that are open, closed, or poised for activation. Chromatin domains have various sizes and different extents of stability. These variations allow for phenomena found solely in eukaryotes, such as transcription at various stages of development and epigenetic memory throughout cell division cycles. They also allow for the maintenance of differentiated cellular states, which is crucial to the survival of multicellular organisms (Struhl, 1999).
[3] While the number hasn’t been locked down for humans, it’s estimated to be 42 million for bacteria, providing a floor for what should be expected in a complex organism.
[4] An estimated 25% of the proteome are cell membrane proteins