I.I Addendum: DNA-based data storage
The amount of data generated each year is already outstripping our ability to store it and that gap will only continue to widen, driving needs to explore higher order storage mechanisms. Global data storage capacity of ~7 zettabytes is one-tenth the 64 ZB[1] of data generated in 2020 alone, up from 2 ZB ten years prior. The order of magnitude differential in data creation and storage capacity is explained by the fact a small fraction of data needs to be stored. With that said, that gap is projected to continue widening at 5% a year. Eventually, that’ll run into our planet’s physical limits. By 2040, data is expected to consume all the world’s microchip-grade silicon without a fundamental change to our relatively inefficient digital archiving processes. Files are currently stored on magnetic tape, disk drives, or flash memory. While tape is cheap to make and query, it takes up meaningful space and must be replaced every 10 years, as it physically degrades.
To find a superior data storage mechanism, we need to look no further than our own bodies. DNA is a 1,000 times denser data storage mechanism than the densest hardware humanity has invented – compact solid-state hard drives – and at least 300-fold more durable than the most stable magnetic tapes. As context, it could store a modern data center in test tubes or all the movies ever made in an area smaller than the size of a sugar cube.[2] Additionally, whereas magnetic tapes must be replaced every 10 years, DNA lasts for tens of thousands of years and almost a million years when stored properly. This is appealing given that more than 90% of the data held in servers like Facebook's data centers is never accessed. It just sits on the shelves waiting to be useful.
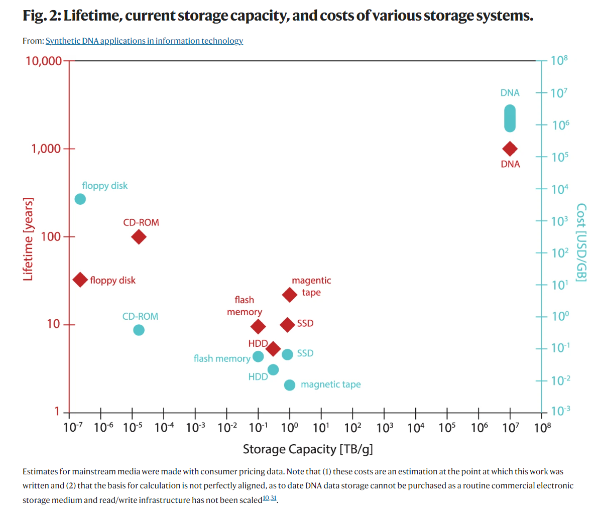
The problem is of course cost and speed. DNA-synthesis companies like Twist Bioscience charge 7-9 cents per base, meaning a single minute of high-quality stereo sound could be stored for nearly $100,000. The speed of our DNA writing technology must also improve by an order of magnitude, as this VC articulates:
For a “DNA drive” to compete with an archival tape drive today, it needs to be able to write ~2Gbits/sec, which is about 2 Gbases/sec. That is the equivalent of ~20 synthetic human genomes/min, or ~10K sHumans/day, if I must coin a unit of DNA synthesis to capture the magnitude of the change. Obviously this is likely to be in the form of either short ssDNA, or possibly medium-length ss- or dsDNA if enzymatic synthesis becomes a factor. If this sDNA were to be used to assemble genomes, it would first have to be assembled into genes, and then into synthetic chromosomes, a non trivial task. While this would be hard, and would to take a great deal of effort and PhD theses, it certainly isn't science fiction.
Querying the data in the DNA drive would require sequencing methods far faster than even our most advanced NGS technologies, which can only read at megabases per second vs hundreds for tape (which is slower than disks or solid-state drives). Moreover, even assuming Ultima’s machine can break the $100 per human genome barrier, that’d mean it’d cost 3 cents per megabase or 12 cents a megabyte. That understates the cost by a lot too, because the human genome is a well studied sequence so a random one without a template would cost more.
The first prototype for DNA storage came from George Church’s lab in 2012. Since then, researchers have encoded all kinds of things in As, Ts, Cs, and Gs: War and Peace, Deep Purple’s “Smoke on the Water,” a galloping horse GIF. Today, researchers across the top universities, government labs, and corporations are racing to develop the technology. DARPA’s Molecular Informatics Program has invested $15M in projects at Harvard, Brown, Illinois, and UW. Mega tech companies like Microsoft, Intel, and Micron are all publicly working on solutions, and it’s reasonable to assume Azure’s cloud competitors in Amazon and Google don’t intend to be left out. Additionally, several sexy startups and biotechs are in the hunt. For instance, Twist’s spending $40M a year to develop a data storage platform.
All told, a lot of progress has been made. The section below surveys the latest work in the field:
Much of the research has involved encoding ever-more data in the DNA ever-more cheaply:
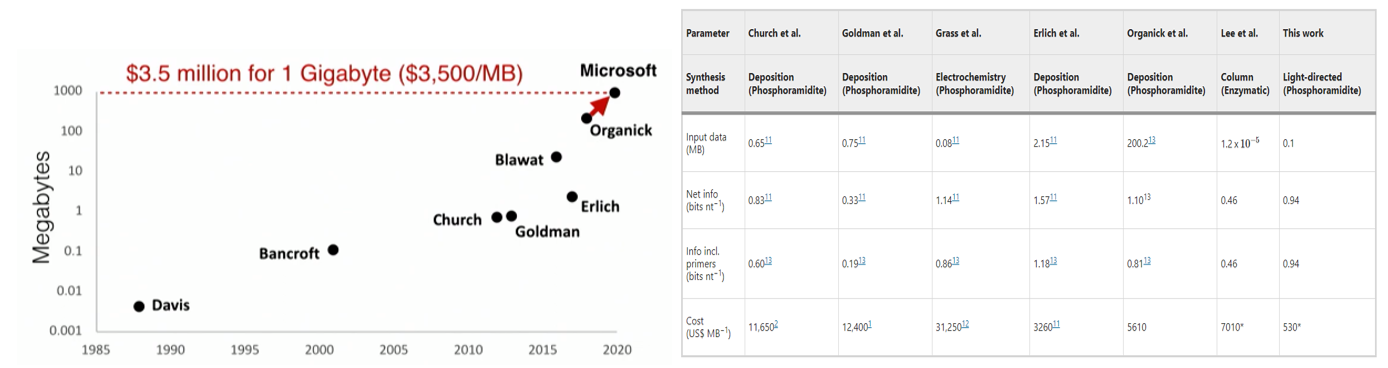
- A research group developed a DNA storage system that uses a far cheaper and higher throughput but correspondingly less accurate synthesis method called massively parallel light-directed synthesis, as opposed to the traditional solid-phase synthesis. They created an error correction algorithm that can handle these high error rates to recover the original data perfectly. Storing a 0.1MB of Mozart sheet music cost $530/MB, as seen in the table above. They also hinted as ways to cut the cost of DNA storage to $1/MB.
- Several of the core founding members of Wyss are collaborating to develop a new enzyme-based approach to write DNA simpler and faster than traditional chemical synthesis. They believe this can reduce the cost of $3,500 / MB by orders of magnitude.
- A top startup in the space called Catalog is building a machine to write a TB of data a day, using 500 trillion molecules of DNA. Its novel approach decouples the process of writing DNA from that of encoding it. Traditional methods map the series of 0s and 1s to the four base pairs. Catalog generates large quantities of a short DNA strands – no longer than 30bp – and then uses enzymatic reactions combine the sequences together. That combinatorial space yields billions of unique molecules upon which it encodes the information. In other words, rather than mapping one bit to one base pair, bits are arranged in multidimensional matrices, and sets of molecules represent their locations in each matrix. This approach avoids having to synthesize new DNA for every new piece of stored information; instead the company can just remix its pre-fabricated DNA molecules.
The firm poached the former head of DNA synthesis at Ginkgo to be the chief science officer and execute the vision. He believes the approach will be cost competitive with tape storage shortly as the firm scales up automation. It plans to launch industrial-scale storage services for tech companies, the entertainment industry, and the federal government. - Microsoft’s improvement on electrochemical synthesis mentioned above is explicitly being developed for use in data storage:
A natural next step is to embed digital logic in the chip to allow individual control of millions of electrode spots to write kilobytes per second of data in DNA. From there, we foresee the technology reaching arrays containing billions of electrodes capable of storing megabytes per second of data in DNA. This will bring DNA data storage performance and cost significantly closer to tape.
Other groups are working on making it easier to query the test tubes of DNA:
- Microsoft developed random access retrieval for large-scale storage so that they don’t have to sequence all the DNA in a test tube when only a small subset of information needs to be returned. They demonstrated their algorithm that greatly reduces the sequencing read coverage necessary for error-free decoding on 35 distinct files of over 200MB of data stored in 13 million oligos.
- Microsoft and UW collaborated to make a fully automated, programmable system that can store and retrieve data in DNA:
The Microsoft-UW team has also created a programmable system that can move droplets of fluid around on a digital microfluidic device dubbed PurpleDrop . The operating system, known as Puddle, can be used to issue commands for a microfluidic system, much as a more conventional operating system like Linux can issue commands for an electronic computing system.
Eventually, a next-generation DNA data storage system could be combined with devices like PurpleDrop and software like Puddle to create a computer environment based on microfluidics instead of electronics. Ceze said that would probably lead to hybrid computer systems that blend the processing power of electronic computing with the data storage density of DNA.
- A team at UW adapted a machine learning technique called “semantic hashing” to design an efficient search-by-image mechanism for querying data stored in DNA. The team developed an encoder to output feature sequences that react chemically more strongly with the feature sequences of similar images. This creates a molecular “fish hook”: when a molecule encoding the desired image is added to the database, molecules representing similar images react with and stick to the queried one. The resulting query-target pairs can then be extracted using magnets. This process makes chemistry do the work otherwise done by computer vision models. Initial experiments were “moderately successful”, hooking 30% of similar files when those files represented 10% of the whole database.
Still others are figuring out how to edit data and conduct computation without ever leaving the realm of chemistry:
- Researchers at Caltech and the University of California at Davis published a paper describing a data processing system that uses self-assembling DNA molecules to run algorithms, enabling computation at the molecular scale, though not necessarily processing large amounts of data.
- In contrast to traditional (passive) DNA storage, schemes for dynamic DNA storage allow access and modification of data without sequencing and/or synthesis. Upon binding to molecular probes, files can be accessed selectively (4) and modified through PCR amplification (5). Introducing or inhibiting binding of molecular probes with existing data barcodes can rename or delete files (6). Information encoded in the hybridization pattern of DNA can be written and erased (7) and can even undergo basic logic operations such as AND and OR (8) using strand displacement. By encoding information in the nicks of naturally occurring DNA [a.k.a. native DNA (9)], data can be modified through ligation or cleavage (10). With image similarities encoded in the binding affinities of DNA query probes and data, similarity searches on databases can be performed through DNA hybridization (11, 12). Although these advances allow information to be directly accessed and edited within the storage medium, they nevertheless demonstrate limited or no capacity for computation in DNA.
- Utilizing our understanding of the kinetics and thermodynamics of DNA strand displacement (13–15), researchers have designed a variety of rationally designed molecular computing devices. These include molecular implementations of logic circuits (16–18), neural networks (19, 20), consensus algorithms (21), dynamical systems including oscillators (22), and cargo-sorting robots (23). Given the achievements of strand displacement systems and their inherent molecular parallelism, DNA computation schemes appear well suited to directly carry out computation on big data stored in DNA.
- UT Austin researchers addressed the fact that processing information on data stored in DNA requires a clunky change of domain from chemical to digital and back by designing a storage system capable of general purpose computation without ever leaving the realm of chemistry:
Inspired by the computational power of “DNA strand displacement,” we augment DNA storage with “in-memory” molecular computation using strand displacement reactions to algorithmically modify data in a parallel manner
Inspired by methods of strand displacement methods of storing information in the positions of nicks in double-stranded DNA (9, 10), our system encodes information in a multistranded DNA complex with a unique pattern of nicks and exposed single-stranded regions (a register). Although storage density is somewhat reduced (approximately a factor of 30; see Discussion), encoding information in nicks still achieves orders of magnitude higher density than magnetic and optical technologies. To manipulate information, an instruction (a set of strands) is applied in parallel to all registers which all share the same sequence space (Fig. 1B). The strand composition of a register changes when the applied instruction strands trigger strand displacement reactions within that register. Nonreacted instruction strands and reaction waste products are washed away via magnetic bead separation to prepare for the next instruction. Each instruction can change every bit on every register, yielding a high level of parallelism. Our experiments routinely manipulated 1010 registers in parallel; DNA storage recovery studies suggest that, in principle, 108 distinct register values can be stored in one such sample
Our work merges DNA storage and DNA computing, setting the foundation of entirely molecular algorithms for parallel manipulation of digital information preserved in DNA.
Read next section: quantifying the cells' attributes and activity (omics)
[1] 64 ZB = 64 billion terabytes or ~17 trillion Blue-ray DVDs worth of data